LAMMPS
LAMMPS is a "Molecular Dynamics Simulator" which supports a wide variety of different force fields. CSC does not have a general purpose installation of LAMMPS, as each user typically needs a more or less customized version. Please read below how to create yours.
Available
- Puhti: Instructions and CMake files available for building in
/appl/soft/chem/lammps/ - Mahti: Instructions and CMake files available for building in
/appl/soft/chem/lammps/ - LUMI: Instructions and CMake files available for building in
/appl/local/csc/soft/chem/lammps
Note
Don't use prebuilt binaries, but take a look at the instructions below for configuring and compiling LAMMPS for optimal performance. Don't hesitate to contact CSC Service Desk if you encounter any problems!
License
LAMMPS is an open-source code, distributed freely under the terms of the GNU Public License (GPL).
Usage
- Navigate to
/appl/soft/chem/lammps/on Puhti/Mahti, or/appl/local/csc/soft/chem/lammpson LUMI. - If you can't find a pre-downloaded source code (e.g.
stable_2Aug2023.tar.gz) or the LAMMPS version is not suitable, download it yourself from the LAMMPS home page. - Read the compilation instructions, e.g.
lammps-cpu-instruction.txt. - Select the packages you want to include and compile the software following the instructions.
- Example input and batch scripts are available in the
exampledirectory.
Compile using the fast local disk
Please compile in $TMPDIR on Puhti/Mahti for faster performance and
less load on the shared file system. As the local disk is cleaned
frequently, remember to move your files to your project's /projappl
directory afterwards. Setting -DCMAKE_INSTALL_PREFIX=/projappl/... will
ensure that the files are moved automatically when running make install.
See the provided build instructions for specific details.
GPU versions
To enable GPU support, we recommend building LAMMPS with the Kokkos package. Kokkos is a portable programming model that allows running on both Nvidia and AMD GPUs. It is typically also more efficient than the standard GPU package.
When running, it is recommended to set the number of MPI tasks per node to be equal to the number of physical GPUs on the node (GCDs on LUMI). Assigning multiple MPI tasks to the same GPU is usually only faster if some portions of the input script have not been ported to use Kokkos. See the LAMMPS documentation for more details.
Consult these pages for generic instructions about running jobs:
- How to create batch jobs on Puhti
- How to create batch jobs on Mahti.
- How to create batch jobs on LUMI
Performance notes
The following diagram compares the performance and scaling of LAMMPS on CPUs and GPUs on Mahti and LUMI. The system contains 16M atoms.
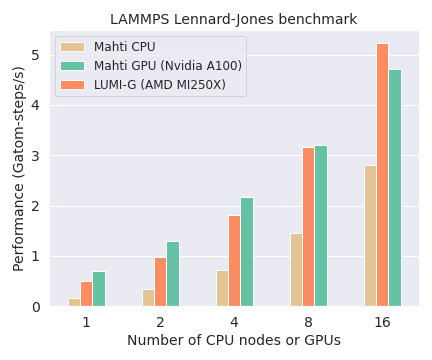
- LAMMPS peformance is measured here in Gatom-steps/s, i.e. how many billion atoms can be propagated one time step each second.
- Large systems (millions of atoms) are able to utilize multiple GPUs efficiently. The example system of 16M atoms scales well to several GPU nodes and is about 10 times faster compared to running on an equal amount of CPU nodes.
- Smaller systems are best run on CPUs or a single GPU (or by sharing one GPU among multiple independent trajectories using multi-replica simulations, see below).
High-throughput computing with LAMMPS
LAMMPS offers comprehensive support for executing loops and multiple
independent simulations using a single input file. The -partition
command-line switch enables running these concurrently within a
single Slurm job step, thus accelerating the computations while keeping the
load on the batch queue system minimal as excessive calls of srun or sbatch
are avoided. An example batch script using the -partition option is provided
for Puhti below.
#!/bin/bash
#SBATCH --account=<project>
#SBATCH --partition=large
#SBATCH --time=00:15:00
#SBATCH --nodes=3
#SBATCH --ntasks-per-node=40
#SBATCH --mem-per-cpu=100
module purge
module load gcc/11.3.0 openmpi/4.1.4 fftw/3.3.10-mpi-omp
export PATH="/path/to/your/lammps/installation/bin:${PATH}"
export OMP_NUM_THREADS=1
srun lmp -in loop.lammps -partition 24x5
The above example runs an umbrella sampling simulation of ethanol adsorption on
a NaCl surface. The simulation consists of 24 iterations where the ethanol
molecule is gradually pulled closer to the surface. These 24 iterations are all
run concurrently using 5 MPI tasks each, which is specified in the batch script
as -partition 24x5. The number of processors must add up to the amount
requested, in this case 3 full Puhti nodes (120 cores). In general, the
partitions do not have to be of equal size, but one could for example specify
-partition 3x30 20 10 for 3 partitions of 30 cores, one of 20 cores and one
of 10 cores (3 Puhti nodes). This does of course not make sense for jobs where
the subtasks are virtually identical, such as here.
If the -partition switch is used one needs to replace the usual index and
loop variable styles used in the input of sequential simulations. The
corresponding styles compatible with multi-partition jobs are world,
universe and uloop. For further details, see the LAMMPS documentation on
running multiple simulations from one input script,
the partition switch and
variable styles compatible with multi-partition jobs.
A batch script for running the same system by sharing a single LUMI GCD (half a GPU) among all 24 replicas could look like:
#!/bin/bash
#SBATCH --account=<project>
#SBATCH --partition=small-g
#SBATCH --time=00:15:00
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=24
#SBATCH --gpus-per-node=1
module load PrgEnv-amd craype-x86-trento craype-accel-amd-gfx90a rocm cray-fftw
export PATH="/path/to/your/lammps/installation/bin:${PATH}"
export MPICH_GPU_SUPPORT_ENABLED=1
export OMP_NUM_THREADS=1
export OMP_PROC_BIND=spread
export OMP_PLACES=threads
srun lmp -in loop.lammps -k on g 1 -sf kk -pk kokkos -partition 24x1
References
The following CPC paper is the canonical reference to use for citing LAMMPS. It gives an overview of the code including its parallel algorithms, design features, performance, and brief highlights of many of its materials modeling capabilities. If you wish, you can also mention the URL of the LAMMPS website in your paper, namely https://www.lammps.org.
- LAMMPS - a flexible simulation tool for particle-based materials modeling at the atomic, meso, and continuum scales, A. P. Thompson, H. M. Aktulga, R. Berger, D. S. Bolintineanu, W. M. Brown, P. S. Crozier, P. J. in 't Veld, A. Kohlmeyer, S. G. Moore, T. D. Nguyen, R. Shan, M. J. Stevens, J. Tranchida, C. Trott, S. J. Plimpton, Comp Phys Comm, 271 (2022) 10817.
References to other methods used in LAMMPS can be found here.