GROMACS
GROMACS is a very efficient engine to perform molecular dynamics simulations and energy minimization particularly of proteins. However, it can also be used to model polymers, membranes and e.g. coarse-grained systems. It also comes with plenty of analysis scripts.
Available
| Version | Available modules | Notes |
|---|---|---|
| 2020.5 | gromacs/2020.5 |
|
| 2020.7 | gromacs/2020.7 |
|
| 2021.4 | gromacs/2021.4-plumed |
Module with Plumed available |
| 2021.5 | gromacs/2021.5gromacs/2021.5-cuda |
GPU-enabled module available |
| 2021.6 | gromacs/2021.6 |
|
| 2022.2 | gromacs/2022.2gromacs/2022.2-cuda |
GPU-enabled module available |
| 2022.3 | gromacs/2022.3gromacs/2022.3-cuda |
GPU-enabled module available |
| 2022.4 | gromacs/2022.4gromacs/2022.4-cuda |
GPU-enabled module available |
| 2023.2 | gromacs/2023.2 |
|
| 2023.3 | gromacs/2023.3 |
|
| 2024.0 | gromacs/2024 |
|
| 2024.1 | gromacs/2024.1 |
|
| 2024.2 | gromacs/2024.2 |
|
| 2024.3 | gromacs/2024.3 |
| Version | Available modules | Notes |
|---|---|---|
| 2020.4 | gromacs/2020.4-plumed |
Module with Plumed available |
| 2020.5 | gromacs/2020.5 |
|
| 2021.3 | gromacs/2021.3 |
|
| 2021.4 | gromacs/2021.4-plumed |
Module with Plumed available |
| 2021.5 | gromacs/2021.5 |
|
| 2022 | gromacs/2022gromacs/2022-cp2k |
Module with CP2K available for QM/MM |
| 2022.1 | gromacs/2022.1gromacs/2022.1-cp2k |
Module with CP2K available for QM/MM |
| 2022.2 | gromacs/2022.2gromacs/2022.2-cuda |
GPU-enabled module available |
| 2022.3 | gromacs/2022.3gromacs/2022.3-cuda |
GPU-enabled module available |
| 2022.4 | gromacs/2022.4gromacs/2022.4-cuda |
GPU-enabled module available |
| 2023.1 | gromacs/2023.1 |
|
| 2023.2 | gromacs/2023.2 |
|
| 2023.3 | gromacs/2023.3 |
|
| 2024.0 | gromacs/2024 |
|
| 2024.1 | gromacs/2024.1 |
|
| 2024.2 | gromacs/2024.2 |
|
| 2024.3 | gromacs/2024.3 |
| Version | Available modules | Notes |
|---|---|---|
| 2023.3 | gromacs/2023.3gromacs/2023.3-gpu |
GPU-enabled module available |
| 2024.2 | gromacs/2024.2gromacs/2024.2-gpugromacs/2024.2-heffte |
GPU-enabled module available Module with heFFTe available for GPU PME decomposition |
| 2024.3 | gromacs/2024.3gromacs/2024.3-gpugromacs/2024.3-heffte |
GPU-enabled module available Module with heFFTe available for GPU PME decomposition |
- Puhti and Mahti have also
gromacs-env/<year>modules for loading the recommended latest minor version from each year (replace<year>accordingly). - To access modules on LUMI, first load the CSC module tree into use with
module use /appl/local/csc/modulefiles - If you want to use command-line Plumed tools, load the Plumed module.
Info
We only provide the MPI version gmx_mpi, but it can be used for grompp,
editconf etc. similarly to the serial version. Instead of gmx grompp,
give gmx_mpi grompp.
License
GROMACS is a free software available under LGPL, version 2.1.
Usage
Initialize recommended version of GROMACS on Puhti or Mahti like this:
Use module spider to locate other versions. To load these modules, you need
to first load required dependencies, which are shown with
module spider gromacs/<version>. To access CSC's GROMACS modules on LUMI,
remember to first run module use /appl/local/csc/modulefiles.
Note
Please use the -maxh flag for mdrun. Setting this equal to or slightly
less than the requested time limit (in hours) will ensure that there's time
for your simulation to write a final checkpoint and end gracefully before
the scheduler terminates the job. If left unspecified, there's a chance
that the job will crash the node(s) it is running on. For general guidance
on managing long simulations, see the
GROMACS manual.
Notes about performance
Note
Please minimize unnecessary disk I/O – never run simulations using
mdrun -v (the verbose flag)!
It is important to setup the simulations properly to use resources efficiently.
The most important aspects to consider (in addition to avoiding -v) are:
- If you run in parallel, make a scaling test for each system – don't use more cores/GPUs than is efficient. Scaling depends on many aspects of your system and used algorithms, not just size.
- Use a recent version – there has been significant speedup and bug fixes over the years. If you switch the major version, remember to check that the results are comparable.
- For large jobs, use full nodes (multiples of 40 cores on Puhti or multiples of 128 cores on Mahti), see examples below.
- Performance on GPUs depends on many factors and what calculations you offload. Please consult the excellent ENCCS online materials for a general overview, or the GROMACS on LUMI workshop materials for how to run efficiently on LUMI-G.
- On LUMI-G it is important to make sure CPUs are bound to the correct GPUs to minimize communication overhead. See examples below and LUMI Docs for more information.
For a more complete description, consult the mdrun performance checklist on the GROMACS page.
Puhti
#!/bin/bash
#SBATCH --time=00:15:00
#SBATCH --partition=small
#SBATCH --ntasks=1
#SBATCH --account=<project>
##SBATCH --mail-type=END # uncomment to get mail
# this script runs a 1 core gromacs job, requesting 15 minutes time
module purge
module load gromacs-env
export OMP_NUM_THREADS=1
srun gmx_mpi mdrun -s topol -maxh 0.2
#!/bin/bash
#SBATCH --time=00:15:00
#SBATCH --partition=large
#SBATCH --ntasks-per-node=40
#SBATCH --nodes=2
#SBATCH --account=<project>
##SBATCH --mail-type=END # uncomment to get mail
# this script runs an 80 core (2 full nodes) gromacs job, requesting 15 minutes time
module purge
module load gromacs-env
export OMP_NUM_THREADS=1
srun gmx_mpi mdrun -s topol -maxh 0.2 -dlb yes
Note
To avoid multinode parallel jobs spreading over more nodes than
necessary, don't use the --ntasks flag, but specify --nodes and
--ntasks-per-node=40 to get full nodes. This minimizes communication
overhead and fragmentation of node reservations.
#!/bin/bash
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=10
#SBATCH --gres=gpu:v100:1
#SBATCH --time=00:15:00
#SBATCH --partition=gpu
#SBATCH --account=<project>
##SBATCH --mail-type=END # uncomment to get mail
module purge
module load gromacs-env/2022-gpu
export OMP_NUM_THREADS=${SLURM_CPUS_PER_TASK}
srun gmx_mpi mdrun -s topol -maxh 0.2 -dlb yes
# additional flags, like these, may be useful - test!
# srun gmx_mpi mdrun -pin on -pme gpu -pmefft gpu -nb gpu -bonded gpu -update gpu -nstlist 200 -s topol -dlb yes
Note
Please make sure that using one GPU (and up to 10 CPU cores) is faster than using one full node of CPU cores according to our usage policy. Otherwise, don't use GPUs on Puhti.
Mahti
#!/bin/bash
#SBATCH --time=00:15:00
#SBATCH --partition=medium
#SBATCH --ntasks-per-node=128
#SBATCH --nodes=2
#SBATCH --account=<project>
##SBATCH --mail-type=END # uncomment to get mail
# this script runs a 256 core (2 full nodes, no hyperthreading) gromacs
# job, requesting 15 minutes time
module purge
module load gromacs-env
export OMP_NUM_THREADS=1
srun gmx_mpi mdrun -s topol -maxh 0.2 -dlb yes
#!/bin/bash
#SBATCH --time=00:15:00
#SBATCH --partition=medium
#SBATCH --ntasks-per-node=64
#SBATCH --cpus-per-task=2
#SBATCH --nodes=2
#SBATCH --account=<project>
##SBATCH --mail-type=END # uncomment to get mail
# this script runs a 256 core (2 full nodes, no hyperthreading) gromacs
# job, requesting 15 minutes time and 64 tasks per node, each with 2 OpenMP
# threads
module purge
module load gromacs-env
export OMP_NUM_THREADS=${SLURM_CPUS_PER_TASK}
srun gmx_mpi mdrun -s topol -maxh 0.2 -dlb yes
LUMI
#!/bin/bash
#SBATCH --partition=small-g
#SBATCH --account=<project>
#SBATCH --time=00:15:00
#SBATCH --nodes=1
#SBATCH --gpus-per-node=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=7
module use /appl/local/csc/modulefiles
module load gromacs/2024.3-gpu
export OMP_NUM_THREADS=${SLURM_CPUS_PER_TASK}
srun gmx_mpi mdrun -s topol -nb gpu -bonded gpu -pme gpu -update gpu -maxh 0.2
#!/bin/bash
#SBATCH --partition=standard-g
#SBATCH --account=<project>
#SBATCH --time=00:15:00
#SBATCH --nodes=1
#SBATCH --gpus-per-node=8
#SBATCH --ntasks-per-node=8
module use /appl/local/csc/modulefiles
module load gromacs/2024.3-gpu
export OMP_NUM_THREADS=7
export MPICH_GPU_SUPPORT_ENABLED=1
export GMX_ENABLE_DIRECT_GPU_COMM=1
export GMX_FORCE_GPU_AWARE_MPI=1
cat << EOF > select_gpu
#!/bin/bash
export ROCR_VISIBLE_DEVICES=\$SLURM_LOCALID
exec \$*
EOF
chmod +x ./select_gpu
CPU_BIND="mask_cpu:fe000000000000,fe00000000000000"
CPU_BIND="${CPU_BIND},fe0000,fe000000"
CPU_BIND="${CPU_BIND},fe,fe00"
CPU_BIND="${CPU_BIND},fe00000000,fe0000000000"
srun --cpu-bind=${CPU_BIND} ./select_gpu gmx_mpi mdrun -s topol -nb gpu -bonded gpu -pme gpu -update gpu -npme 1 -maxh 0.2
Terminology
Each GPU on LUMI is composed of two AMD Graphics Compute Dies (GCD). Since there are four GPUs per node, and Slurm interprets each GCD as a separate GPU, you can reserve up to 8 "GPUs" per node. See more details in LUMI Docs.
Notes about binding and multi-GPU simulations on LUMI
Only certain CPU cores are directly linked to a specific GPU on LUMI, so to maximize multi-GPU performance, it is important to ensure that CPU cores are bound to the GPUs accordingly. The full GPU node example above takes care of this, and also excludes the first core of each group of 8 cores linked to a given GCD. These are reserved for the operating system to reduce noise, meaning that there are only 56 cores available per node. This is also why we run 7 threads per MPI rank, not 8.
Note
Please note that CPU-GPU binding only works when reserving full nodes by
running in the standard-g partition or by using the --exclusive flag.
See more details in LUMI Docs:
LUMI-G hardware,
LUMI-G examples,
GPU binding
Instead of communicating between GPUs through the CPU, direct GPU communication will also bring significant performance benefits when running on multiple GPUs. Enabling this requires setting the following environment variables in your batch script (see also the full GPU node example above):
export MPICH_GPU_SUPPORT_ENABLED=1
export GMX_ENABLE_DIRECT_GPU_COMM=1
export GMX_FORCE_GPU_AWARE_MPI=1
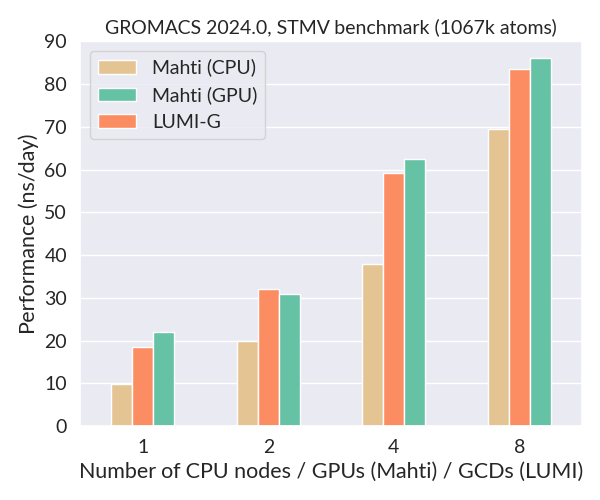
Below is a comparison of the performance of GROMACS 2024.3 on Mahti (CPUs and GPUs) and LUMI-G using the STMV benchmark (1067k atoms, 2 fs timestep). This is a large system which scales very well also on GPUs. The performance of a single LUMI GCD (half a GPU) is about the same as a full Nvidia A100 GPU on Mahti, and much better than a single 128-core CPU node. Importantly, the availability of GPU nodes on LUMI is massive compared to Mahti (2978 vs. 24).
Small systems and high-throughput simulations
While medium-sized and large systems (few 100k to 1M+ atoms) can typically utilize multiple GPUs well, small systems (less than 100k atoms) are often best run on just a single GCD. A good way to further increase the GPU utilization and efficiency of small simulations is to share one GCD between multiple independent trajectories. This can be accomplished using the built-in multidir feature of GROMACS. For more details about GPU-sharing and aggregate sampling, see our tutorial on high-throughput simulations with GROMACS.
GPU PME decomposition
The scalability of huge systems with several million atoms may be limited by
single GPU PME. To significantly improve scalability, decomposition of PME work
to multiple GPUs is possible in modules suffixed with -heffte which have been
linked to the heFFTe library. Add the
following exports to your batch script:
The number of PME ranks to use depends on the specific case, but 1 or 2 per GPU
node should be a reasonable starting point. So for 16 LUMI-G nodes, try
-npme 16 or -npme 32.
Visualization and analysis
GROMACS trajectory files and data can be visualized, for example, with the following programs:
- VMD visualization program for large biomolecular systems
- Grace plotting data produced with GROMACS tools
- MDAnalysis Python library to analyze
trajectories from MD simulations
- Not available at CSC, but can be easily installed by the user in a containerized Conda environment with Tykky
- PyMOL molecular modeling system (not available at CSC)
More are listed in the GROMACS manual. In addition, GROMACS itself includes numerous post-processing utilities for analyzing trajectories. See the command-line reference for details.
Running heavy/long analyses
Visualization of large trajectories, as well as certain GROMACS tool scripts,
can be computationally very demanding and should never be run on the login
nodes (see usage policy). Instead, please run
such workloads in an
interactive session. Since we
only provide the MPI-version of GROMACS, you need to prepend your gmx_mpi
command with orterun -n 1, e.g.:
sinteractive --account <project>
module load gromacs-env
orterun -n 1 gmx_mpi msd -n index -s topol -f traj
As most GROMACS analysis utilities, such as the msd tool above, can only be
run in serial, they might take quite long for large trajectories. In such cases
it may be more convenient to run the tools as serial batch jobs. If
the command you want to run requires interaction (e.g. to select which parts of
your system to include in the analysis), you may pass these in a batch job for
example like this:
# Three consecutive selections (options 2, 2 and 0), you need to know these beforehand
echo "2 2 0" | gmx_mpi trjconv -f traj -s topol -o trajout -pbc cluster -center
Note that you may use the interactive partition (time limit 7 days) also in
batch jobs if the 3 day time limit of small is not enough. The 14-day
longrun partition has a very low priority and using it will often require
substantial queueing. Another viable option is to use the
persistent compute node shell
available through the web interfaces, which will keep running even if you close
your browser or lose internet connection.
References
Cite your work with the following references:
- S. Páll, A. Zhmurov, P. Bauer, M. J. Abraham, M. Lundborg, A. Gray, B. Hess, E. Lindahl. Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J. Chem. Phys. 153 (2020) pp. 134110.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess, E. Lindahl. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1 (2015) pp. 19-25.
- S. Páll, M. J. Abraham, C. Kutzner, B. Hess, E. Lindahl. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. In S. Markidis & E. Laure (Eds.), Solving Software Challenges for Exascale 8759 (2015) pp. 3-27.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess, and E. Lindahl. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29 (2013) pp. 845-54.
- B. Hess and C. Kutzner and D. van der Spoel and E. Lindahl. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4 (2008) pp. 435-447.
- D. van der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen. GROMACS: Fast, Flexible and Free. J. Comp. Chem. 26 (2005) pp. 1701-1719.
- E. Lindahl and B. Hess and D. van der Spoel. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 7 (2001) pp. 306-317.
- H. J. C. Berendsen, D. van der Spoel and R. van Drunen. GROMACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 91 (1995) pp. 43-56.
See your simulation log file for more detailed references for methods applied in your setup.
More information
- GROMACS home page and documentation
- mdrun performance checklist
- Materials at the BioExcel website
- GROMACS community forum
- Poster about the performance of GROMACS on LUMI
- Training materials:
- Tutorials:
- Example
.tprfiles for testing: