Amber
Amber is a molecular dynamics package including a number of additional tools for more sophisticated analysis and in particular NMR structure refinement.
Available
| Version | Available modules |
|---|---|
| 20 | amber/20amber/20-cuda |
| 22 | amber/22amber/22-cuda |
| 24 | amber/24amber/24-cuda |
| Version | Available modules |
|---|---|
| 20 | amber/20amber/20-cuda |
| 22 | amber/22amber/22-cuda |
| 24 | amber/24amber/24-cuda |
| Version | Available modules |
|---|---|
| 24 | amber/24-cpuamber/24-gpu |
License
Amber can be used on CSC servers by all not-for-profit institute and university researchers irrespective of nationality or location. Look for the academic license text here.
Usage
See available versions and how to load Amber by running:
On LUMI, you need to add the CSC modules to your module path before running the above command:
The module load command will set $AMBERHOME and put the AmberTools binaries
in the path. Run Amber production jobs in the batch queues, see below. Very
light system preparation (serial AmberTools jobs lasting a few seconds and
using barely any memory) can be done on the login node as well. Heavier
analyses can be run e.g. in an
interactive compute session.
Molecular dynamics jobs are best run with pmemd.cuda as they are much faster
on GPUs than on CPUs. Please note that using pmemd.cuda requires a module
with the -cuda extension. Similarly on LUMI one should use pmemd.hip (or
pmemd.hip.MPI for multi-GPU simulations), which requires loading a module
with the -gpu extension.
Run only GPU aware binaries in the GPU partitions. If you're unsure, check with
seff <slurm_jobid> that GPUs were used and that the job was significantly
faster than without GPUs.
Python modules
Python scripts distributed with AmberTools are only available in the Amber22 modules on Puhti/Mahti. However, since AmberTools is also distributed through Conda, you can easily create a containerized environment containing these scripts yourself using Tykky or the LUMI container wrapper.
Example batch scripts for Puhti and Mahti
#!/bin/bash
#SBATCH --time=00:10:00
#SBATCH --partition=gputest
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --account=<project>
#SBATCH --gres=gpu:v100:1
# Our tests show that for medium-sized systems the most efficient setup is
# one GPU card and one CPU core.
module purge
module load gcc/9.4.0 openmpi/4.1.4
module load amber/22-cuda
srun pmemd.cuda -O -i mdin -r restrt -x mdcrd -o mdout
#!/bin/bash
#SBATCH --time=00:10:00
#SBATCH --partition=test
#SBATCH --ntasks=1
#SBATCH --account=<project>
# The non-GPU aware binaries, e.g. AmberTools, can be run as batch jobs in
# the following way:
module purge
module load gcc/9.4.0 openmpi/4.1.4
module load amber/22
srun paramfit -i Job_Control.in -p prmtop -c mdcrd -q QM_data.dat
#!/bin/bash
#SBATCH --time=00:10:00
#SBATCH --partition=gputest
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --account=<project>
#SBATCH --gres=gpu:a100:1
module purge
module load gcc/9.4.0 openmpi/4.1.2
module load amber/22-cuda
srun pmemd.cuda -O -i mdin.GPU -o mdout.GPU -p Cellulose.prmtop -c Cellulose.inpcrd
Note
pmemd.cuda (and pmemd.hip on LUMI) are way faster than pmemd.MPI, so
use a CPU-version only in case you cannot use the GPU-version. If Amber
performance is not fast enough, consider using GROMACS, which
is typically able to scale further (i.e. make use of more CPU and/or GPU
resources). Consider also whether you really need speed or just a lot of
sampling. Accelerated sampling can also be achieved through ensemble
simulations, where multiple independent trajectories (e.g. the same system
equilibrated from different initial velocities) are run at the same time.
For more details, see the section on
high-throughput simulations with Amber.
If you want to use more than one GPU, perform scaling tests to verify that
the jobs really become faster and use a binary with .cuda.MPI or
.hip.MPI extension. The rule of thumb is that when you double the
resources, the job duration should decrease at least 1.5-fold. For overall
performance info, consult the
Amber benchmark scaling details.
Typically, the best efficiency is achieved with 1 GPU.
Example GPU batch scripts for LUMI
Amber can be loaded into use on LUMI with:
#!/bin/bash
#SBATCH --partition=small-g
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --gpus-per-node=1
#SBATCH --time=01:00:00
#SBATCH --account=<project>
module use /appl/local/csc/modulefiles
module load amber/24-gpu
srun pmemd.hip -O -i mdin.GPU -o mdout.GPU -p Cellulose.prmtop -c Cellulose.inpcrd
#!/bin/bash
#SBATCH --partition=standard-g
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=8
#SBATCH --gpus-per-node=8
#SBATCH --time=01:00:00
#SBATCH --account=<project>
module use /appl/local/csc/modulefiles
module load amber/24-gpu
export MPICH_GPU_SUPPORT_ENABLED=1
cat << EOF > select_gpu
#!/bin/bash
export ROCR_VISIBLE_DEVICES=\$SLURM_LOCALID
exec \$*
EOF
chmod +x ./select_gpu
CPU_BIND="mask_cpu:fe000000000000,fe00000000000000"
CPU_BIND="${CPU_BIND},fe0000,fe000000"
CPU_BIND="${CPU_BIND},fe,fe00"
CPU_BIND="${CPU_BIND},fe00000000,fe0000000000"
srun --cpu-bind=$CPU_BIND ./select_gpu pmemd.hip.MPI -O -i mdin.GPU -o mdout.GPU -p Cellulose.prmtop -c Cellulose.inpcrd
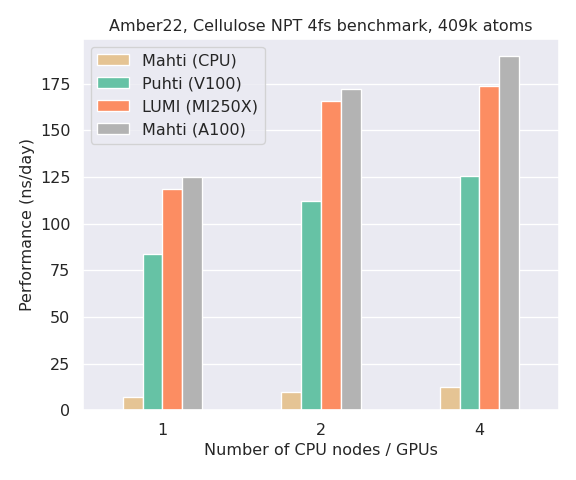
A performance comparison of Amber on CPUs (Mahti) and GPUs (Puhti, Mahti, LUMI) is shown in the bar plot below. Notice how the performance of a single GPU on all systems is an order of magnitude better than a full Mahti CPU node (128 cores). Note also that Amber is typically not able to scale efficiently to multiple GPUs unless you have a very large system (>1 million particles).
GPU binding on LUMI
Running on multiple GPUs on LUMI will benefit from GPU binding. In the
example above, a bitmask is used to bind CPU cores to optimal (linked) GPUs
as well as exclude the first CPU core in each group of 8 cores (these are
reserved for the operating system and thus not available for computing).
For background and further instructions, see the
LUMI documentation.
Note that CPU/GPU binding is only possible when reserving full nodes
(standard-g or --exclusive).
Generic batch script examples for LUMI-G and LUMI-C are available in the LUMI documentation.
Interactive jobs
Sometimes it is more convenient to run small jobs, like system preparations, interactively. Interactive batch jobs prevent excessive load on the login node and should be used in these kinds of cases. You can request a shell on a compute node from the Puhti/Mahti web interface, from the command line with sinteractive, or manually with:
Once you have been allocated resources (you might need to wait), you can run
e.g. the paramfit task directly with:
High-throughput computing with Amber
Similar to GROMACS multidir, Amber has a built-in "multi-pmemd" functionality, which allows you to run multiple MD simulations within a single Slurm allocation. This is an efficient option in cases where you want to run many similar, but independent, simulations. Typical use cases are enhanced sampling methods such as replica exchange MD. Also, since Amber simulations do not typically scale that well to multiple GPUs, multi-simulations can be used as a straightforward method to accelerate sampling by launching several differently initialized copies of your system, all running simultaneously on a single GCD each. If your system is very small and hence unable to utilize the full capacity of a GCD, it might even make sense to run multiple replicas on the same GCD to maximize efficiency.
Note
GPU resources on Puhti and Mahti are scarce, so we recommend running large-scale multi-pmemd simulations only on LUMI. LUMI-G has a massive GPU capacity available, which is also more affordable in terms of BUs compared to Puhti and Mahti.
An example multi-pmemd batch script for LUMI-G is provided below.
#!/bin/bash
#SBATCH --partition=standard-g
#SBATCH --nodes=2
#SBATCH --ntasks-per-node=8
#SBATCH --gpus-per-node=8
#SBATCH --time=01:00:00
#SBATCH --account=<project>
module use /appl/local/csc/modulefiles
module load amber/24-gpu
export MPICH_GPU_SUPPORT_ENABLED=1
cat << EOF > select_gpu
#!/bin/bash
export ROCR_VISIBLE_DEVICES=\$SLURM_LOCALID
exec \$*
EOF
chmod +x ./select_gpu
CPU_BIND="mask_cpu:fe000000000000,fe00000000000000"
CPU_BIND="${CPU_BIND},fe0000,fe000000"
CPU_BIND="${CPU_BIND},fe,fe00"
CPU_BIND="${CPU_BIND},fe00000000,fe0000000000"
srun --cpu-bind=$CPU_BIND ./select_gpu pmemd.hip.MPI -ng 16 -groupfile groupfile
In this example, 16 copies of a system are run concurrently within a single
Amber job, each using 1 GCD. 2 nodes are requested in total as each node on
LUMI-G contains 8 GCDs (4 GPUs). The input, output, topology and coordinate
files for the respective simulations are defined in a so-called groupfile:
-O -i mdin.GPU -o mdout000.GPU -p system000.prmtop -c system000.inpcrd
-O -i mdin.GPU -o mdout001.GPU -p system001.prmtop -c system001.inpcrd
-O -i mdin.GPU -o mdout002.GPU -p system002.prmtop -c system002.inpcrd
-O -i mdin.GPU -o mdout003.GPU -p system003.prmtop -c system003.inpcrd
-O -i mdin.GPU -o mdout004.GPU -p system004.prmtop -c system004.inpcrd
-O -i mdin.GPU -o mdout005.GPU -p system005.prmtop -c system005.inpcrd
-O -i mdin.GPU -o mdout006.GPU -p system006.prmtop -c system006.inpcrd
-O -i mdin.GPU -o mdout007.GPU -p system007.prmtop -c system007.inpcrd
-O -i mdin.GPU -o mdout008.GPU -p system008.prmtop -c system008.inpcrd
-O -i mdin.GPU -o mdout009.GPU -p system009.prmtop -c system009.inpcrd
-O -i mdin.GPU -o mdout010.GPU -p system010.prmtop -c system010.inpcrd
-O -i mdin.GPU -o mdout011.GPU -p system011.prmtop -c system011.inpcrd
-O -i mdin.GPU -o mdout012.GPU -p system012.prmtop -c system012.inpcrd
-O -i mdin.GPU -o mdout013.GPU -p system013.prmtop -c system013.inpcrd
-O -i mdin.GPU -o mdout014.GPU -p system014.prmtop -c system014.inpcrd
-O -i mdin.GPU -o mdout015.GPU -p system015.prmtop -c system015.inpcrd
See the Amber manual for further details on multi-pmemd.
References
When citing Amber or AmberTools, please use the following:
D.A. Case, H.M. Aktulga, K. Belfon, I.Y. Ben-Shalom, J.T. Berryman, S.R. Brozell, D.S. Cerutti, T.E. Cheatham, III, G.A. Cisneros, V.W.D. Cruzeiro, T.A. Darden, N. Forouzesh, M. Ghazimirsaeed, G. Giambaşu, T. Giese, M.K. Gilson, H. Gohlke, A.W. Goetz, J. Harris, Z. Huang, S. Izadi, S.A. Izmailov, K. Kasavajhala, M.C. Kaymak, A. Kovalenko, T. Kurtzman, T.S. Lee, P. Li, Z. Li, C. Lin, J. Liu, T. Luchko, R. Luo, M. Machado, M. Manathunga, K.M. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, K.A. O'Hearn, A. Onufriev, F. Pan, S. Pantano, A. Rahnamoun, D.R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, A. Shajan, J. Shen, C.L. Simmerling, N.R. Skrynnikov, J. Smith, J. Swails, R.C. Walker, J. Wang, J. Wang, X. Wu, Y. Wu, Y. Xiong, Y. Xue, D.M. York, C. Zhao, Q. Zhu, and P.A. Kollman (2024), Amber 2024, University of California, San Francisco.
More Information
The Amber home page has an extensive manual and useful tutorials.